Table of Contents
Rendering UI
Motivation
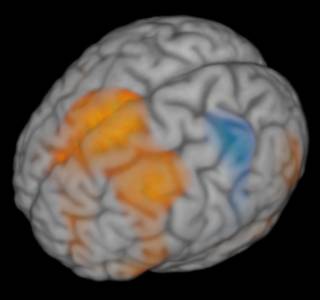
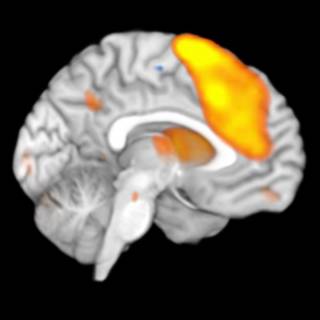
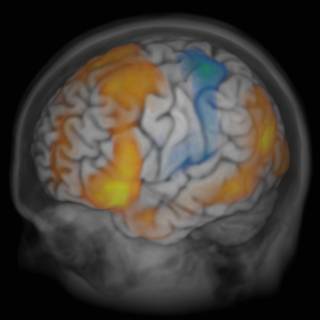
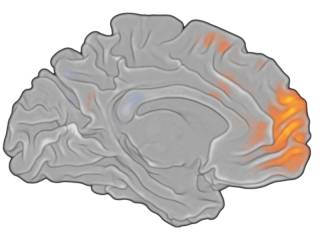
Whereas slice-displays can give a very “close-to-the-data” look at results of a whole-brain mapping, it sometimes is more desirable to visualize an entire 3D space with one image. One way to do this is by using mesh-based surfaces containing a projection of statistical information. However, the extent in the third dimension (orthogonal to the surface) is not easy to visualize in (and grasp from) such images. Another look at the data is to render the entire dataset slice by slice in a semi-transparent fashion. Here are four examples (click to zoom):
Layout
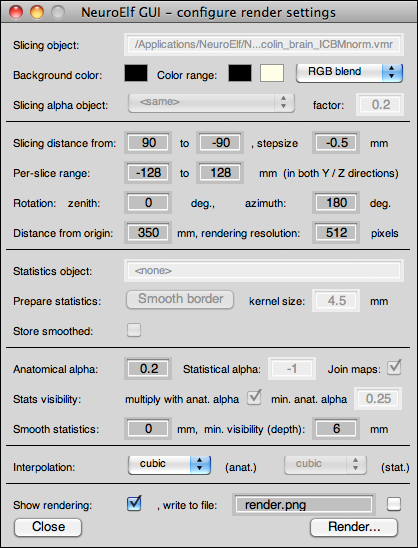
The Rendering UI is available via the Visualization → Render menu item as well as the rendering button (below the surface pane button), and it looks like this:
The dialog has the following controls and properties:
- read-only edit box showing the anatomical dataset used for rendering (whatever object is selected while the dialog is invoked)
- colored buttons for the background as well as the lower and upper boundary values for the anatomical dataset, followed by a dropdown box defining the color-blending mechanism
- drop-down box to chose an additional (secondary) anatomical dataset used for alpha (transparency) information (e.g. to make certain parts of the volume semi-transparent), followed by the alpha-level factor for this secondary dataset (edit box)
- slice stepping controls (from, to, step size) which determine which planes in the 3D space will be sampled (smaller step sizes increase quality for high-res images)
- planar bounding box controls (Y/Z from and to) which determine the coordinate space per sampled slice
- rotation controls (same logic as with surface pane, which can be used as a “preview” to find a good viewing angle)
- distance control which determines the amount of perspective-based shearing
- resolution control which sets the output image size (square)
- statistical map preprocessing controls, allowing to smooth the edges of a VMP (e.g. after clustering) to obtain a higher quality “inlay” to a brain (and resulting in less edges)
- anatomical alpha-level (relative to black/white sampled color of brain; unused if a secondary dataset is selected for the alpha channel sampling)
- statistical alpha-level (negative values produce soft edges, as lower thresholds will be scaled between 0 and 1 internally already)
- join maps, controls whether for multiple selected maps the colors are to represent a mixed color (joined) or the “top-most” color (in order of VMP container)
- statistics visibility restriction settings (multiplication with anatomical alpha checkbox and minimum anatomical alpha requirement)
- statistics smoothing, this controls whether, regardless of edge smoothing, the sampled slices will be smoothed (does not affect actual map)
- minimal visibility (through how much space of brain/skull should activations be visible at least?)
- interpolation method choice dropdowns (separately for anatomical and statistical datasets)
- show continuously updated version of rendering
- target filename and saving checkbox
- UI close button
- Render button
Configuration examples
The images above have been created as follows (using the same t-map for all three images):
Common preparation
For all images, the contrast map has been restricted to the brain of the visualization subject (colin_brain_ICBMnorm.vmr). Next, the map has been “border-smoothed” using an 8mm kernel to remove sharp edges around the brain border. The anatomical alpha was set to 0.2. The visibility (min. depth) has been set to 3.5mm, and a slice step size of 0.5mm has been set.
Full brain and activation
The resolution was set to 1024 pixels, and the angles were 60 degrees zenith and 150 degrees azimuth.
Medial view on left hemisphere
A copy of the brain dataset was created from which the right hemisphere was removed. This copy was slightly smoothed to account for some rough edges at the cutting plane.
Brain and skull view
The full head dataset (colin_ICBMnorm.vmr) was selected in the main UI before invoking the rendering UI, and the background of the brain VMR was filled with a low value (30), and then selected as the secondary alpha channel, which makes all non-brain voxels appear with a very high transparency.
Back-projected surface
The left-hemisphere (160k vertices version) was back-projected to a 0.5mm VMR resolution VMR, border-smoothed, then filled up with a gray intensity value, and then rendered with an activation map overlaid.