This is an old revision of the document!
Table of Contents
GSR data analysis
Motivation
For many experimental tasks (e.g. subjects follwing a set of specific but differing instructions), it is often helpful to collect secondary physiological measurements, such as galvanic skin-response (GSR) over time (both as a manipulation check but also as potential candidates for mediation analysis). The recorded data often requires manual clean-up and inspection, and this page should give you some pointers as to how to conduct some of these steps.
Overview
The following steps are covered:
- reading in data files
- re-sampling data
- (pre-) filtering data
- onset information extraction/synchronization from/with GSR data
- obtaining peak-to-peak measures
Reading in data files
Currently, the following input formats are supported:
- ACQ (up until version ⇐ 3.9.7)
- generic MAT file
- TXT (use
object = xff('*.ntt');or object =xff(filename, 'ntt');to read!)
To load the data from an ACQ file, use the following syntax:
- GSR_data_readacq.m
% use file selector of xff class gsr = xff('*.acq', 'Please select the GSR-ACQ file to work with...'); % the actual data will then either be in data = gsr.RawData(CHANNEL, :)'; % or in data = gsr.Channel(CHANNEL).Data;
To use a variable in a MAT file, simply use the load MATFILE call in Matlab.
Or if you require to load the data from a text-based (e.g. log) file, you can use the following code:
- GSR_data_readtxt.m
% use file selector of xff class (you might have to change the file type % in the selector from ''*.txt'' to ''*.*'' in case the extension is not TXT! gsr = xff('*.ntt', 'Please select the text-based GSR file to work will...'); % the data is then available as data = gsr.Data(:, CHANNEL);
Re-sampling data
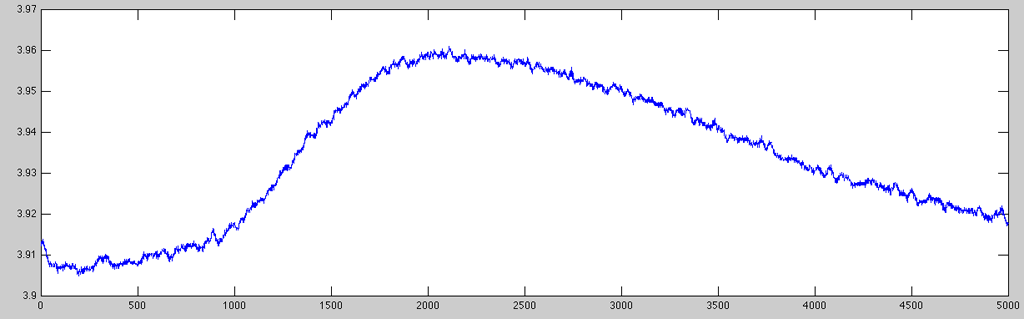
Very often, the raw data acquisition sampling rate is much higher than the required rate to perform any meaningful analyses on GSR data (e.g. peak-to-peak measurements). Here is an example of ten seconds of raw data acquired at 500Hz (5,000 data points):
- GSR_rawplot.m
% getting the data data = gsr.Channel(1).AmplitudeScale .* gsr.RawData(1, :)'; % plotting from second 504 through 514 plot(data(252000:256999));
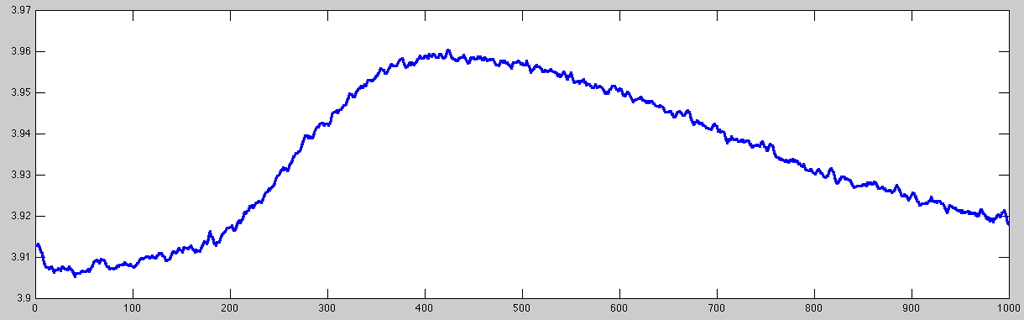
As you can see, the very small signal variations are normally quite uninteresting (as the skin response only very sluggishly follows the stimulation/emotional state of the subject). Hence, it is usually a good idea to resample the signal to a more suitable resolution. To keep some of the finer details (and being able to determine latency values with a reasonable resolution), for this example I resampled the data to 100Hz using the resampleaa function (which works a little better than using just downsample):
- GSR_resampledplot.m
% resampling the data rdata = resampleaa(data, 5); % plotting same time index with stronger line set(plot(rdata(50400:51399)), 'LineWidth', 2);
(Pre-) Filtering data
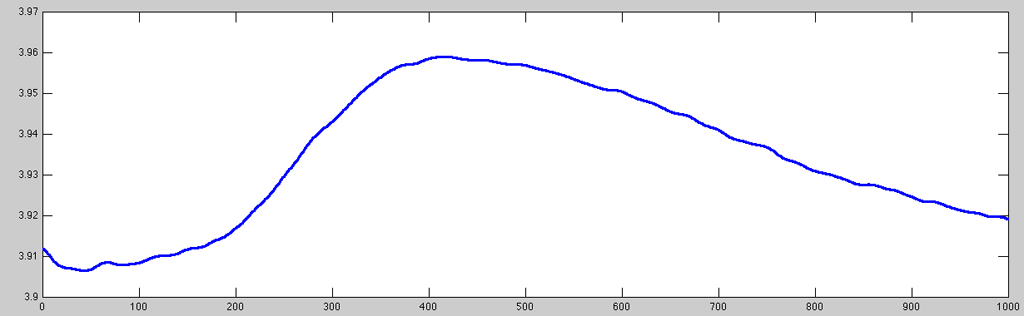
While a lot of the very high-frequency noise is (naturally) gone after the resampling, there is still a lot of signal variation that is (very likely) unrelated to any task/stimulation the subject could have reacted to. Further the signal pre-processing, I then applied a difference (first discreet derivative, diff)-filter that removes artifacts in signals which are unlikely to be in the desired spectrum using the prefilter function:
- GSR_prefilter.m
% simply pass to prefilter fdata = prefilter(rdata); % plotting same time index with stronger line set(plot(fdata(50400:51399)), 'LineWidth', 2);
Now the data looks reasonably clean to undergo further processing (e.g. peak-to-peak analysis).
Onset information
In our first example, the onsets are available are channels in the ACQ object we use. The condition (type of stimulus) is encoded in three channels (2 through 4) and the within-trial stimulus distinction (fixation, instruction, pre-stimulus, stimulus, rating) is given by channels 5 through 9. These channels either carry a zero value (condition/stimulus not present) or a value greater than 0 (present).
To obtain a list of onsets, we first find the onsets of all conditions and then lookup those values in the vectors required for the different conditions (in our case three), whereas the onset of interest is coded as channel 8 (stimulus):
- GSR_acq_stimonsets.m
% all stimuli onsets (use 1 + to cope for the fact that the % first value in diff rather codes the second sample in the data! all_onsets = 1 + find(diff(gsr.RawData(8, :)') > 0); % now split into three conditions neu = all_onsets(gsr.RawData(2, all_onsets) > 0); neg = all_onsets(gsr.RawData(3, all_onsets) > 0); rea = all_onsets(gsr.RawData(4, all_onsets) > 0); % cope for resampling from 500Hz -> 100Hz !! neu = round(0.2 .* neu); neg = round(0.2 .* neg); rea = round(0.2 .* rea);
As window length for the inspection of the GSR data, we choose 15 seconds, so in the resolution of the resampled data that means 1,500 samples.
Peak-to-peak analysis
Once we are happy with the onsets, the plotcurves function can be used to show each window from onset to offset and then determine the peak-to-peak response:
% create required options argument for plotcurves
gsrchannel = 1;
neucurves = cell(numel(neu), 2);
negcurves = cell(numel(neg), 2);
reacurves = cell(numel(rea), 2);
for sc = 1:numel(neu)
neucurves{sc, 1} = sprintf('LookNeu %02d at t=%.3fs', sc, 0.01 * neu(sc));
neucurves{sc, 2} = [gsrchannel, neu(sc), neu(sc) + 1499];
end
for sc = 1:numel(neg)
negcurves{sc, 1} = sprintf('LookNeg %02d at t=%.3fs', sc, 0.01 * neg(sc));
negcurves{sc, 2} = [gsrchannel, neg(sc), neg(sc) + 1499];
end
for sc = 1:numel(rea)
reacurves{sc, 1} = sprintf('ReaNeg %02d at t=%.3fs', sc, 0.01 * rea(sc));
reacurves{sc, 2} = [gsrchannel, rea(sc), rea(sc) + 1499];
end
% combine curves
curves = cat(1, neucurves, negcurves, reacurves);
% define sets
sets = { ...
'LookNeu', (1:numel(neu))'; ...
'LookNeg', (numel(neu)+1:numel(neu)+numel(neg))'; ...
'ReaNeg', (numel(neu)+numel(neg)+1:size(curves,1))'};
% use plot curves
plotcurves(gsr, struct('curves', {curves}, 'sets', {sets}));