This is an old revision of the document!
Table of Contents
NeuroElf main UI
Motivation
While the toolbox is primarily written for advanced users who know there way around Matlab and are able to make full use of the toolbox functionality, the need for a rudimentary graphical user interface (GUI) arose soon after introducing the toolbox to my fellow lab members.
neuroelf_gui is the response to that need and might, in future versions, show extented capabilities.
GUI layout
The GUI comes in two modes, full and minimized (whereas minimized removes some UI elements from the figure so as to save some space for other windows).
To bring up the GUI, simply issue the
% bring up NeuroElf GUI
neuroelf_gui
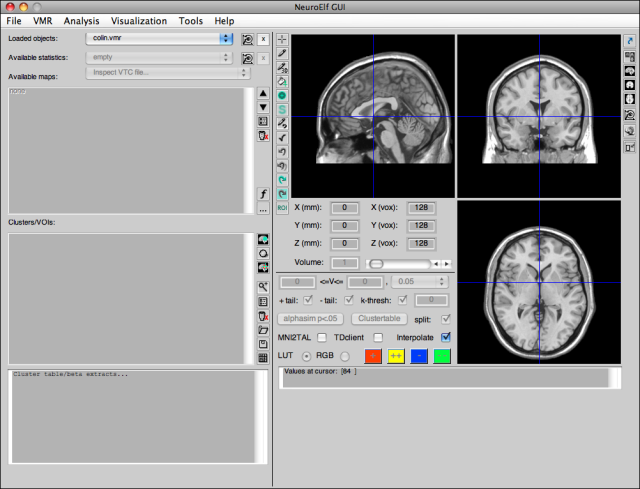
call. It will take a couple of seconds (especially if the xfigure class needs to be run-time compiled by Matlab on its first call), after which the following UI figure will appear:
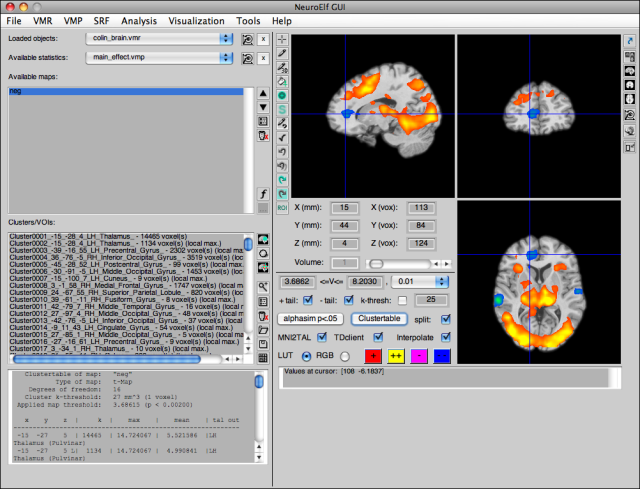
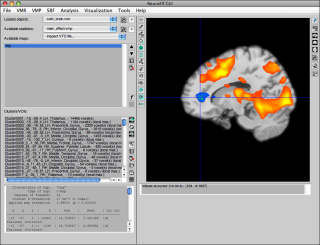
After loading or creating some statistical map (and, in this case, the Colin brain VMR), the display has more buttons un-grayed:
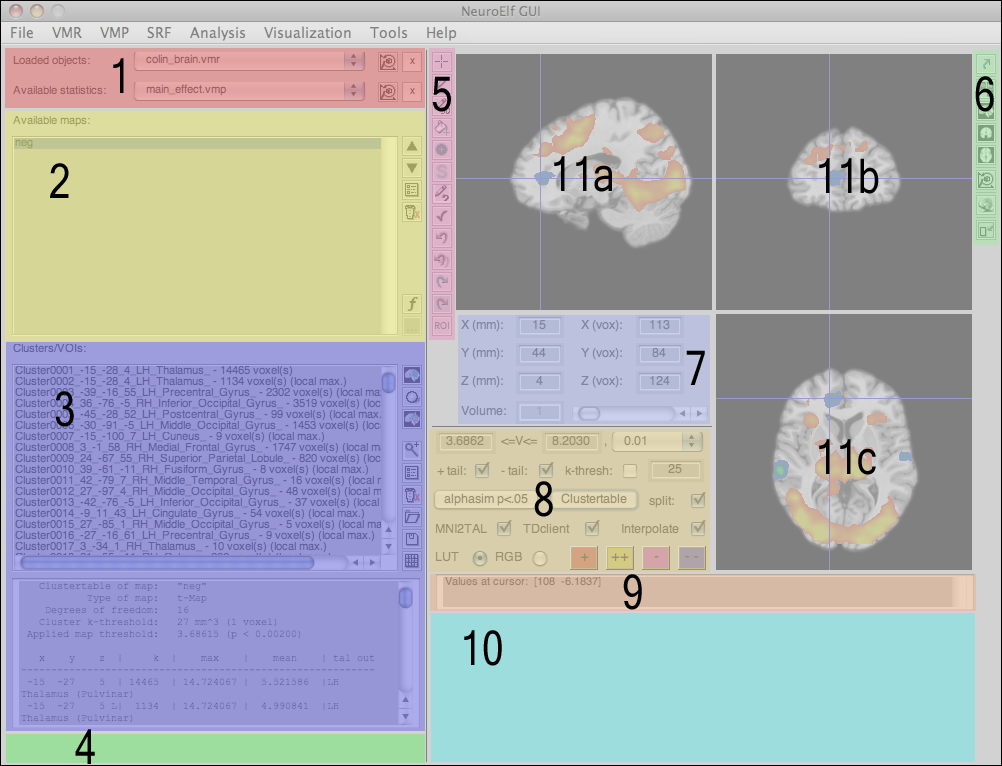
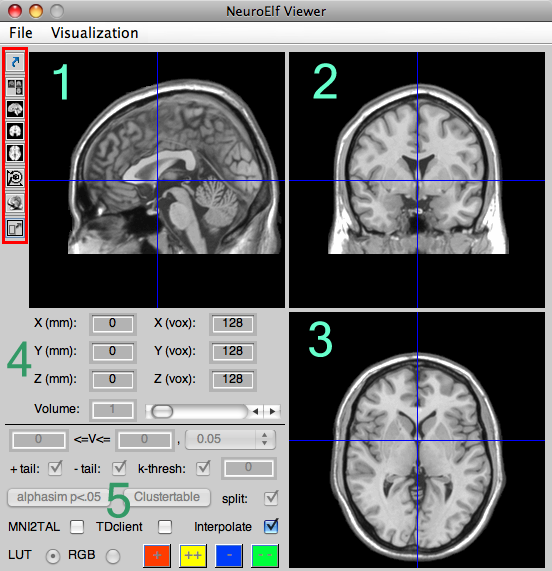
Here is a full-resolution version with numbered areas which are explained below:
The numbered sections in turn mean:
- dataset selection (among loaded objects)
- statistical map selection (multi-select capable)
- configured clusters/regions of interest and output of cluster table (only for VMP/SMP format at this time)
- progress bar area (only visible if a long-running task is executed)
- dataset manipulation tools (only HDR/NII/VMR at this time)
- view (page) selection buttons and global transformation configuration
- cursor position in space (TAL/MNI coordinate and BV system coordinate, as well as volume selector)
- configuration of statistical map (only VMP format at this time)
- output of values at current cursor position (anatomical and statistical values of all selected maps)
- optional time course display if selected dataset is functional and multi-volume (FMR, 4D NII, and VTC)
- canonical slicing of anatomical (or functional + optional statistical) dataset(s)
- a) sagittal view
- b) coronal view
- c) axial/transversal view
Dataset selection
The dropdown controls allow to select any of the loaded datasets as the currently active dataset (both for anatomical and statistical datasets). Currently supported are
- as anatomical datasets
- FMR (BrainVoyager's Functional MR format)
- HDR/NII (Analyze/NIftI format, incl. 4D NIftI files)
- HEAD (AFNI's BRIK/HEAD format, anatomical auto-detect)
- VMR (BrainVoyager's Volumetric MR format)
- VTC (BrainVoyager's Volumetric TimeCourse format)
- as statistical datasets
- GLM (BrainVoyager's General Linear Model format)
- HEAD (AFNI's BRIK/HEAD format, statistical auto-detect)
- VMP (BrainVoyager's Volumentic Map format)
Please note: Statistical maps from SPM must currently be imported into BrainVoyager's VMP format for thresholded map display!
In addition to the selection, for each dataset a transformation quaternion (origin) can be set separately, in case datasets from different programs are not initially compatible (3D transformation button). Once a dataset is no longer required in the list, it can be removed from the dropdown (using the x-button).
Statistical map selection
Once an object is loaded suitable for statistical map overlay, the maps therein can be displayed, also supporting a multi-selection of maps. Additionally, the following actions are supported:
- moving maps within their container (HEAD and VMP format, arrow up and down buttons)
- setting map properties (VMP format only, properties button)
- deleting maps from their container (HEAD and VMP format, trashcan button)
- computing a formula on the available map data (VMP format only, f-button, see vmp.ComputeFormula for more info)
- propagating the map condition selection for other subjects (GLM format, ellipse-button)
Configured clusters and table output
Below the map area is the listbox that contains the currently configured clusters (comparable to BrainVoyager's ROI dialog). Each time the Clustertable button in the stats area is used, the clusters will be overwritten, so make sure to save any desired/altered list of clusters before using the Clustertable function again!
The buttons next to the cluster list are (from top to bottom)
- mark cluster (sets voxel values in VMR project to specific code)
- restrict cluster (restricts the cluster to either a box or sphere around the first/peak voxel)
- find nearest peak (computes distances between current position and cluster peaks and selects closest peak/cluster)
- zoom on/off (enables a zoomed view on that cluster (potentially useful for screenshots with all three slicing views)
- cluster properties (edit cluster properties)
- delete selected clusters
- load cluster configuration
- save cluster configuration
- extract betas from currently selected GLM for selected clusters (auto-extract is enabled by default)
Below the list of clusters is a multi-line text edit field which contains
- a text version of the clustertable after pressing on the Clustertable button (potentially also showing the nearest gray-matter Talairach label for each coordinate)
- a table of extracted betas (also showing each subject's name and the label of the cluster from which the data was drawn)
Progress bar
For some longer-running tasks (e.g. alphasim or robust regression of contrasts), a progress bar is shown and updated regularly. This allows the UI to remain mostly function (browsing, shorter tasks) while a longer task is completing in the back-ground.
Dataset manipulation buttons
From top to bottom, these buttons are associated with the following actions/settings:
- browsing mode (drawing disabled)
- 2D drawing mode (drawing within the slice that is clicked into)
- 3D drawing mode (drawing across slices, spherical or cubic around the clicked coordinate)
- floodfill (3D flood-filling of voxels that match the selected criteria)
- marker expansion (expanding the marked selection in a dilation fassion)
- smoothing (the marked selection is smoothed to remove rough edges)
- undo drawing en/disable (drawing takes back values from the buffer)
- accept changes (copy current version into undo buffer)
- undo (copy undo buffer into current version)
- reload (reload dataset from disk)
- reload marked (reload dataset from disk and mask with marked color)
- reload unmarked (reload dataset from disk and inverse-mask with marked color)
View / Page selection
The main UI has three different views:
The view/page selection buttons, from top to bottom, mean:
- undocking (create a secondary, satellite figure containing the current view, allowing to investigate multiple maps/datasets, potentially at the same coordinate via linked browsing)
- switch to three-slice view (page 1, default on startup)
- switch to sagittal-zoomed view (page 2, SAG)
- switch to coronal-zoomed view (page 2, COR)
- switch to axial-zoomed view (page 2, TRA)
- set global spatial transformation parameters (in case data comes from a different program)
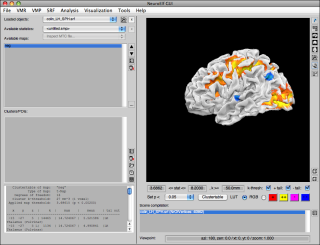
- switch to surface rendering view (page 3)
- minimize/maximize window
Upon clicking this last, bottom-most button, the window will convert to minimized view (if it is in full mode, that is):
The elements on the left side of the figure (file/map selector, cluster and table output) will be hidden from view to free up some space for other windows. The following 5 sections remain:
- sagittal view
- coronal view
- axial/transversal view
- current position
- statistics configuration
Cursor position in space
The six spatial coordinate edit fields show and allow to set the current position (cursor/crosshair) in either TAL/MNI spatial convention (coordinate origin is the center of each slice at coordinate [0, 0, 0]) or BrainVoyager convention (coordinate origin is located in the upper, frontal and right “corner”, so that the TAL-based center is at BV coordinate [128, 128, 128]). Currently only integer coordinates are supported (but the zoomed view allows for interpolated/non-integer coordinates to be shown of course).
Additionally, for four-dimensional datasets, a volume selector and edit field allows to change the currently shown volume.
Configuration of statistical maps
The first row of controls allows to set minimum and maximum thresholds for coloring the statistical map (any value below the minimum will not be shown, values above the maximum will be shown in the color of the maximum), and if the type and degrees of freedom of a statistical map is well-configured, one of the pre-defined probability threshold values can be selected as well (which automatically honors the number of tails selected below for one- or two-tailed statistical thresholding).
In the second row, the positive and negative tails of the configured map can be en- and disabled separately, and also the cluster size threshold can be set and dis/enabled.
The further controls are
- the
alphasim p<.05button uses the current map as a template (values non-equal to zero) and runs alphasim with that template as a mask (using the correct spatial resolution) to determine the required cluster size threshold for the more typical uncorrected p-thresholds (results printed in the Matlab console window). - the
Clustertablebutton will call the vmp.ClusterTable method for the currently select map (only for single map selection) and update the configured clusters list as well as the table output field. The checkbox right of the button dis/enables the splitting of larger clusters into sub-clusters, for which the peaks will be reported separately if enabled. - the
MNI2TALcheckbox controls whether, for any created cluster table, coordinates are converted from MNI into TAL space/convention (by applying themni2tal.mfunction written by M. Brett). - the
TDclientcheckbox controls whether, for any created cluster table, coordinates are passed on to tdclient, so that for each peak the nearest gray-matter label is looked up - the
Interpolatecheckbox controls whether statistical maps are interpolated into the spatial resolution of the UI (1x1x1 mm) or sampled with nearest-neighbor sampling - the
LUTandRGBradio buttons alter the statistical-value to color coding from lookup table (LUT) to RGB color range - if the color coding is set to RGB, the colors for positive and negative minimum and maximum values (thresholds) can be configured using the four colored buttons
Output of values
This box simply contains the sampled value at the current (cursor/crosshair) position, both for the selected anatomical and any selected statistical map.
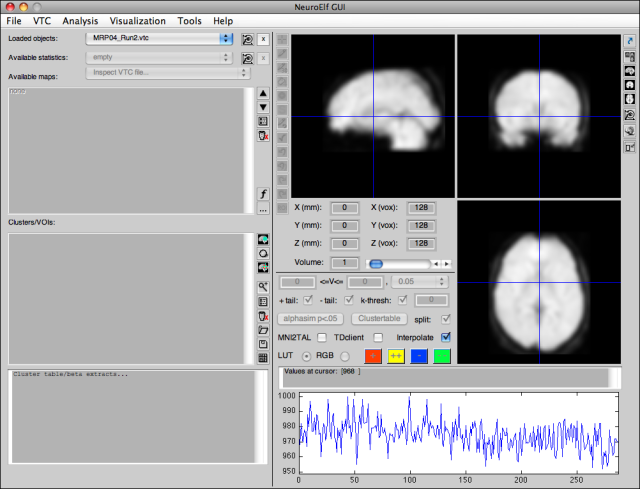
Time course display
For four-dimension datasets, this area displays the time course at the current (cursor/crosshair) position. A click into the timecourse will also set the volume selector to that position (which works more interactively than using the volume slider, as this requires the mouse button to be released!)
Slice display
In both the three-slices view and the zoomed one-slice view, any click into one of the slices will set the cursor to the clicked position within the slice. This allows for relatively simply browsing of the dataset. The position will keep updating while the mouse button remains pressed (and the mouse moved).
To reset the slice display to its original position (central coordinate), press the r key on the keyboard.
Surface display
For the surface display, the following viewpoint modifications are supported with the mouse (while the mouse button is pressed within the surface area and remains pressed throughout the mouse motion):
- without any modifier key pressed, the view will rotate (along the Z axes for left/right mouse movement → longitude/azimuth, and along a perpendicular spatial axis → latitude/zenith)
- with the
SHIFTkey pressed, the view will translate (shift) with the mouse movement - with the
ALTkey pressed, the view will zoom (up → smaller, down → larger; intuitively as if the object were moved away/toward the user along with the mouse)
To reset the surface display to its original view (azi: 180, zen: 0, zoom: 1), press the r key on the keyboard.
Keyboard commands
When no control is currently selected (click between controls, into the background of the figure), the following keyboard commands are supported:
- cursor left/right: for slice-based view change the current (cursor/crosshair) position in X direction +/- 1; for surface view rotate azimuth/longitude by +/- 5
- cursor up/down: for slice-based view change the current (cursor/crosshair) position in Z direction +/- 1; for surface view rotate zenith/latitude by +/- 5
SHIFT+ cursor left/right: for surface view translate view by +/- 5SHIFT+ cursor up/down: for slice-based view change the current (cursor/crosshair) position in Y direction +/- 1; for surface view translate view by +/- 5ALT+ cursor up/down: for slice-based view change alpha-blending in 0.1 increments up/downCTRL+ cursor up/down: for surface view zoom by factor 1.01/0.99akey: toggle crosshair display on/offckey: toggle cluster-check for current map on/off (same as checkbox, which is automatically updated)ikey: toggle interpolation on/off (same as checkbox, which is automatically updated)jkey: switch between multi-map join modes: color-sum-blending and later-map-winslkey: toggle cluster-split checkbox on/offmkey: switch between interpolation methods: trilinear and cubicnkey: negate alpha-blending option, possibly auto-level for multi-map selectionokey: toggle between radiological and neurological display conventionrkey: reset viewskey: toggle between minimized and full viewtkey: switch through three-slices view and the three single-slice zoomed views (order: 3-slice, sagittal, coronal, axial/transversal)ukey: toggle undo drawing on/offxkey: switch cluster auto-beta-extraction on/offzkey: toggle brain zooming flag (for cluster-based zoom)1key: toggle positive statistical tail on/off2key: toggle negative statistical tail on/off
File menu
The File menu allows to load any xff-supported object, but only those displayable by the GUI will be added to the lists of selectable objects (FMR, HDR/NII, HEAD, SMP, SRF, VMP, VMR, VTC)
Additionally, any file loaded via the GUI will (if appropriate) be added to the list of recently loaded objects, so that in an upcoming session, formerly loaded files can be easily re-loaded.
Under the Options submenu, the following entries are available:
- Stats colors (LUT)
- a list of files bundled with NeuroElf to alter the colortable for statistical maps
- Edit LUT palette - this calls up
colorpickerfor the currently selected LUT object - Linked browsing - if several voxel-views (at least one satellite figure with an anatomical dataset) is open, this will set the coordinate in all open windows upon clicking into any of those figures
- Echo function call/methods - if checked, this will cause NeuroElf GUI to print a line of code into the Matlab prompt window that can be used as a template for writing your own scripts
Analysis menu
Currently this menu has only one working entry:
- Contrast manager - this will bring up the Contrast manager UI
Visualization menu
Currently this menu has only one entry:
- Create montage images - this will bring up the Image montage UI
Tools menu
The tools menu gives access to the following actions:
- SPM to BV conversion
- SPM.mat to PRT conversion (spmmat2prt.m)
- SPM.mat to SDM conversion (spmmat2sdm)
- Quality assessment