Table of Contents
NeuroElf - MDM VOI condition averages
Motivation
While hypothesis testing usually is done via formulating a Null-hypothesis (e.g. no systematic effect of differential processing of conditions), for which then a probability is given that the data could have been obtained if the Null-hypothesis were true (and a small enough p-value leading to rejection of this Null-hypothesis in favor of an alternative explanation: the difference in condition leads to a meaningful effect), false-positives can (and do!) still occur frequently. As one additional piece of evidence, it can be of great value to assess whether the region of interest (functionally or anatomically defined, or even just a peak voxel) shows a typical response profile in case the effect is observed in a classical stimulus-driven experiment (different stimuli or instructions to stimuli are contrasted).
One hopes to find a response shape (over time) that resembles the canonical response and, if such a shape is observed, one can have additional confidence in the findings (as opposed to observing more or less random fluctuations over time, which simply happened to “subtract” to a “significant” but possibly false-positive finding).
Requirements
To use this feature in NeuroElf, the following files must be available:
- a VOI file must be loaded (using the load-clusters button in the cluster toolbar)
- VTC files of all runs of subjects which are to be included in the average plots
- PRT files describing the onsets at which stimuli of different conditions were presented
- an MDM file referencing those VTC and PRT files (an MDM referencing SDM files will not work, given that the onsets are no longer available!)
- optionally referenced motion parameter files and temporal filtering settings in the
mdm.RunTimeVarsstructure (see the Storing additional information sub-subsection of the xff class overview page)
Invoking the UI
To bring up the condition-based averaging plot, make sure the correct list of clusters is being displayed in the list of VOIs/clusters (the duration for the extraction roughly increases linearly with the number and size of clusters!), then select the Create VOI condition plots entry from the Analysis menu.
Once an MDM is selected and loaded, NeuroElf will examine the protocol files and show a list of conditions found in those files for selection. This is a good time to remove any conditions of no interest (fixation and rating periods, active baseline, etc.) as this will speed up the subsequent computations.
Next, the time courses will be extracted for all VOIs in all VTCs in the MDM file (subject selection is implemented at a later stage). This can take up to several minutes, depending on the number of VOIs and VTCs for which this has to be done.
In case this is required by the settings (motion parameters and/or temporal filtering), design matrices will be created for the runs, so that time courses can be filtered.
Eventually, a new UI appears.
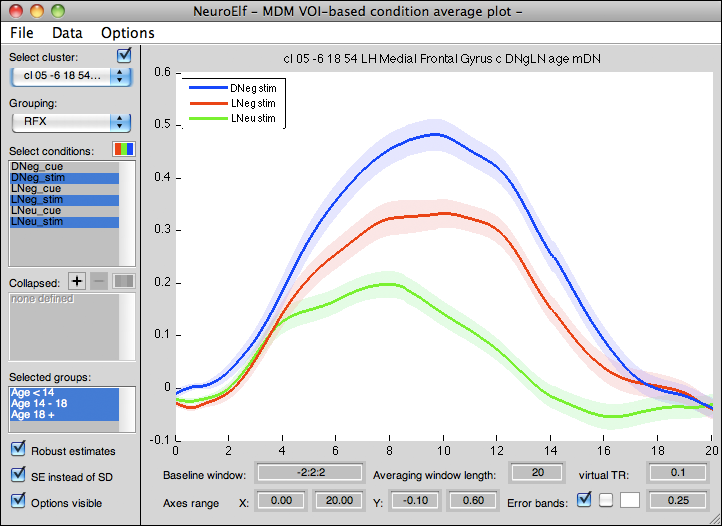
GUI layout
Controls
The following controls are available:
- Select cluster (dropdown): changes the currently shown VOI/cluster
- Update position (checkbox): if checked sets the current position (and cluster, unless reloaded) in the main UI to the cluster peak coordinate
- Grouping (dropdown) with settings FFX, RFX, and RFX (weighted)
- FFX: error bands represent fixed-effects standard deviation (SD) or standard error (SE) of the mean
- RFX: error bands represent random-effects SD or SE of the mean (an intermediate within-subject averaging step precedes any subsequent computation)
- RFX (weighted): this is not yet implemented!! this option shows a random-effects SD or SE, whereas outlier subjects are down-weighed
- Select conditions (multi-selection listbox): the selected conditions will be averaged and plotted (one plotted line/band per selected condition)
- Condition color settings (button): this button allows to re-set the colors for each condition (as they first appear in a referenced PRT file) for visualization purposes
- Collapsed conditions (multi-selection listbox): the configured collapsings will be performed and averaged plots will be shown collapsed (one plotted line/band per selected collapsing)
- Add collapsing (button): takes the currently selected conditions and creates a collapsing configuration (requesting a name), this in effect allows to combine onsets of several conditions at a late stage instead of having to re-create protocol files
- Remove collapsing (button): removes the currently selected collapsings from the list
- Collapsed conditions color settings (button): this button allows to define new colors for each collapsing
- Selected groups (multi-selection listbox): if groups are configured in the
mdm.RunTimeVars.Groupsfield (same syntax as with GLM group definition), sub-groups of subjects can be (de-) selected for visualization - Robust estimates (checkbox): if checked averages will be performed with outliers removed
- SE instead of SD (checkbox): if checked (and error bands are visualized) error bands represent SE of the mean instead of SD of the mean (allowing to relate inferential statistical outcome to plotted curves; please note that this does not allow to draw inferential conclusions on its own!!)
- Options visible (checkbox): if unchecked, all other controls are hidden and the axes is maximized to cover (almost) the entire figure (improved screenshots)
- Baseline window (edit field): expression indicating the (relative-to-onsets) timepoints (in seconds!) that are used to compute the baseline for each extracted window (must be in Matlab's interval notation using three terms:
From:Stepsize:To!), upon entering a new expression, the entire data will be recomputed, which can take a couple of seconds - Averaging window length (edit field): a single number indicating the length of the window being extracted (in seconds!)
- virtual TR (edit field): a single number indicating the virtual resolution (in seconds!) of the visualization (allowing to improve response overlap for non-TR locked designs; very small numbers will increase the computational and plotting effort!)
- Axes range X/Y from/to (edit fields): if the auto-scaling of the axes object is undesirable (e.g. to use the same scaling across several regions/conditions/studies/etc.), the XLim and YLim axes properties can be set via these four edit controls; to revert to auto-scaling, simply enter an invalid number (e.g. any letter)
- Error bands (checkbox): if checked, also visualize error band indicators (see also SE instead of SD setting above)
- Individual response lines (checkbox, unlabeled): if checked, also plot individual responses (or within-subject averages for RFX setting; in the FFX case this can mean that VERY many lines are plotted, slowing down the UI!)
- Background color setting (button): this button can be used to alter the background color of the axes
- Error bands alpha setting (edit field): this number is used to set the alpha (transparency) property of the error bands
Menu
The following functions are available via a menu:
- File
- Save extract datafile… - stores the MDM/VOI content along with the extracted timecourses (and optionally created SDMs) and all settings to a
.MAT-file for later use (which then doesn't require any additional data, just NeuroElf for visualization!) - Save figure screenshot… - calls the private screenshot function of NeuroElf, requesting a filename, saving the figure as an image
- Save axes screenshot… - moves the axes object to a new figure and then calls the private screenshot function of NeuroElf, requesting a filename, saving the axes object incl. the scale and title as an image
- Data
- Set Data in Base Workspace - make the data currently used available in the base workspace
- Options
- Legend position - allows to alter the position of the legend sub-axes to either of the four axes corners
- Subject selection - brings up a listbox selector (
listdlg) with the subject names, allows to exclude individual subjects from the plotter (e.g. to see if a particular subject is an outlier in all conditions, or otherwise shows an abnormal pattern)
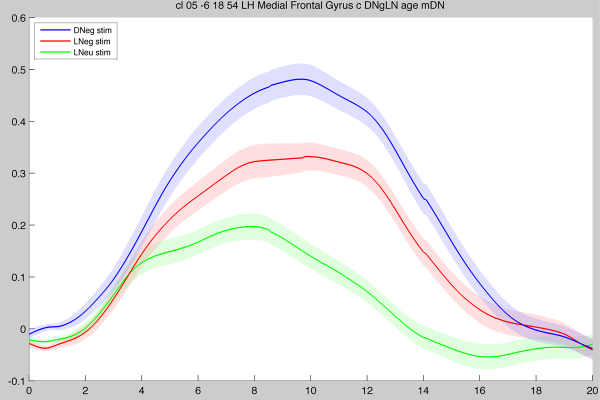
High-quality screenshot example
Using the File → Save axes screenshot… menu item, it is possible to create high-quality plots by choosing the Encapsulated PostScript (.eps) format and then loading the EPS file into a graphics program (e.g. Adobe Photoshop or The GIMP). The above screenshot is then rendered (at any desired resolution), which will give nicer images for publication (please click on the image to zoom and compare!):