Table of Contents
NeuroElf - compute multi-study GLM
Motivation
While looking at the outcome of regressions on single runs (e.g. via using the VTC::CreateGLM and GLM::SingleStudy_tMap methods) can already be instructive, most neuroimaging studies focus on trying to draw conclusions that can be generalized to the entirety of subject population (where the participants in the actual study represent a randomly drawn sample, and one candidate for an appropriate statistical test thus is a random-effects t-map).
This is currently the only menu-based and fully integrated way of performing full-volume-based time-course data regression in NeuroElf, and it is implemented via the MDM::ComputeGLM method. To simplify file selection, NeuroElf comes with its own UI for this task.
Requirements
Please read the page on GLM computation carefully before proceeding, so that you are aware of the options (not all of which are currently implemented in the GUI version, such as PPI settings, etc.).
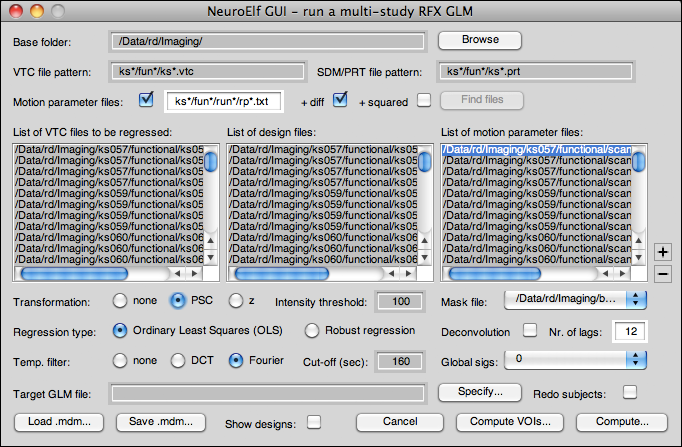
GUI layout
Controls
The following controls are available:
- Base folder (edit field): This shows the location relative to which all file search operations are performed; it is thus imperative to store the functional and design information data in a systematic way across subjects, using the same coding rules and folder structure for each of them!
- Base folder browse (button): Clicking on this button will bring up a folder selection dialog (
uigetdir), and upon successfully selecting a folder, the Base folder edit field will be set to the selected folder location - VTC file pattern (edit field): file pattern identifying all VTC files to be found upon clicking the Find files button (see below)
- SDM/PRT file pattern (edit field): file pattern identifying all design specification files to be found upon clicking the Find files button
- Motion parameter files (checkbox): if checked, a click on the Find files button will also try to locate one motion parameters file per found VTC
- Motion parameter files (edit field, pattern): file pattern identifying the motion parameter files to be found upon clicking the Find files button (if and only if the preceding checkbox is ticked)
- + diff (checkbox): if checked (and motion parameter files are being used) also adds the first derivative of the motion parameters as confounds to the design matrix (see the .motparsd flag)
- + squared (checkbox): if checked (and motion parameter files are being used) also adds the squared (and mean-removed) motion parameters as confounds to the design matrix (see the .motparsq flag)
- Find files (button): after entering and selecting the desired configuration of file names (patterns), clicking this button will use two (or three) calls to findfiles to locate the VTC and PRT (as well as motion parameter) files for the current analysis; if the number and/or names of folders containing the files mismatch, an error message is given, otherwise the filenames are shown in the listboxes underneath
- List of VTC/design/motion parameter files (multi-selection listboxes): “linked” listboxes (selection of files in one listbox will also set the same selection in the other (two) enabled listbox)
- Add VTC+PRT/SDM (plus button): adds a single run's worth of data (single VTC and design specification file) to the list of files
- Remove selection (minus button): deletes the currently selected files from the lists
- Transformation (radio button group) with settings none, PSC, and z:
- Intensity threshold (edit field): this value is passed as .ithresh flag
- Mask file (dropdown): if opened and selected will bring up a file selector for a mask file
- Regression type (radio button group) with settings OLS and robust:
- OLS (Ordinary Least Squares): regression is performed under the single condition of minimizing the sum of squares of the residual using the normal equations where the residual term is orthogonal (sub-space) to the regressors (by simply matrix inversion and multiplication)
- robust: in principle comparable with OLS regression, but iteratively performs re-weighting of datapoints that, by the bisquare weighting function, are considered to be outliers (repeated up to 50 times, until weights converge on stable solution, see fitrobustbisquare_img and its default settings)
- Deconvolution (checkbox): if checked perform a deconvolution (see .ndcregs flag)
- Nr. of lags (edit field): passed as .ndcregs flag if and only if the previous checkbox is ticked (otherwise this flag is set to 0)
- Temp. filter (radio group) with settings none, DCT, and Fourier:
- none: no temporal filtering occurs (no addition to the design matrices)
- DCT: a set of DCT (discrete cosine transform) regressors is added to the design matrices
- Fourier: a Fourier basis set up to a certain frequency is added to the design matrices
- Cut-off (sec) (edit field): temporal filtering cut-off in seconds
- Global sigs (dropdown): this control is not yet fully supported only the numerical values (0, 1, or 2) are passed on as a number via the .globsigs flag
- Target GLM file (edit field): filename for the resulting GLM file (if given, also sets the .loadglm flag to
true) - Specify (button): if clicked opens a file-saving dialog (
uiputfile) and fills the Target GLM file edit field - Redo subjects (checkbox): if an existing filename is specified, the
.loadglmflag will be set totrue(see above), and the user can then choose whether or not to only “add” new subject or to redo all subjects referenced in the current MDM selection - Load .mdm (button): brings up a file request dialog and loads the selected MDM file from disk
- Save .mdm (button): brings up a file-saving dialog and saves the MDM file (including the
.RunTimeVarssettings) to disk - Show designs (checkbox): if checked shows the designs during computation (as plots)
- Cancel (button): closes the UI without further changes
- Compute VOIs… (button): not yet functional, will use the MDM::ComputeVOIGLM method equivalently, dialog is missing a VOI selection tool!
- Compute… (button): pass execution to the MDM::ComputeGLM method, opens the resulting GLM, and then closes the UI