This is an old revision of the document!
Table of Contents
NeuroElf main UI
Motivation
While the toolbox is primarily written for advanced users who know there way around Matlab and are able to make full use of the toolbox functionality, the need for a rudimentary graphical user interface (GUI) arose soon after introducing the toolbox to my fellow lab members.
neuroelf_gui is the response to that need and might, in future versions, show extented capabilities.
GUI layout
The GUI comes in two modes, full and minimized (whereas minimized removes some UI elements from the figure so as to save some space for other windows).
To bring up the GUI, simply issue the
<code matlab>% bring up NeuroElf GUI neuroelf_gui</code
call. It will take a couple of seconds (especially if the xfigure class needs to be run-time compiled by Matlab on its first call), after which the following UI figure will appear:
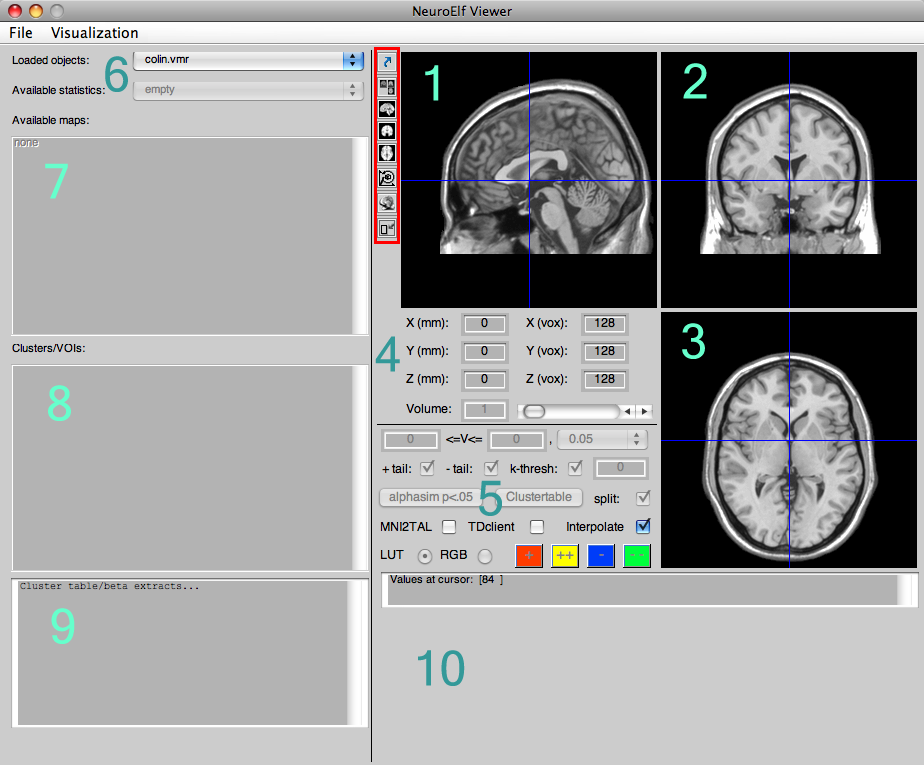
The numbered sections in turn mean:
- sagittal view of the selected anatomical (and/or functional/statistical) dataset(s)
- coronal view of the selected anatomical (and/or functional/statistical) dataset(s)
- axial/transversal view of the selected anatomical (and/or functional/statistical) dataset(s)
- cursor position in space (TAL/MNI coordinate and BV system coordinate, as well as volume selector)
- configuration of statistical map (only VMP format at this time)
- dataset selection (among loaded objects)
- statistical map selection (multi-select capable)
- configured clusters/regions of interest
- output of cluster table (only for VMP format at this time)
- optional time course display if selected dataset is functional and multi-volume (FMR, 4D NII, and VTC)
The buttons in the red-marked box, from top to bottom, mean:
- undocking (create a secondary, satellite figure containing the current view, allowing to investigate multiple maps/datasets, potentially at the same coordinate via linked browsing)
- switch to three-slice view (default on startup)
- switch to sagittal-zoomed view
- switch to coronal-zoomed view
- switch to axial-zoomed view
- set global spatial transformation parameters (in case data comes from a different program)
- switch to surface rendering view
- minimize/maximize window
Upon clicking this last, bottom-most button, the window will convert to minimized view (if it is in full mode, that is):
The elements on the left side of the figure (file/map selector, cluster and table output) will be hidden from view to free up some space for other windows.
File menu
The File menu allows to load any xff-supported object, but only those displayable by the GUI will be added to the lists of selectable objects (FMR, HDR/NII, HEAD, SMP, SRF, VMP, VMR, VTC)
Additionally, any file loaded via the GUI will (if appropriate) be added to the list of recently loaded objects, so that in an upcoming session, formerly loaded files can be easily re-loaded.
Under the Options submenu, the following entries are available:
- Stats colors (LUT)
- a list of files bundled with NeuroElf to alter the colortable for statistical maps
- Edit LUT palette - this calls up
colorpickerfor the currently selected LUT object - Linked browsing - if several voxel-views (at least one satellite figure with an anatomical dataset) is open, this will set the coordinate in all open windows upon clicking into any of those figures
- Echo function call/methods - if checked, this will cause NeuroElf GUI to print a line of code into the Matlab prompt window that can be used as a template for writing your own scripts
Visualization menu
Currently this menu has only one entry:
- Create montage images - this will bring up the NeuroElf - montage UI