This is an old revision of the document!
Table of Contents
heartbeats - ECG / heart-rate data analysis
Motivation
For many tasks, it is often helpful to collect secondary physiological measurements, such as heart-rate over time. The data processing can be rather complicated.
Requirements
The raw signal (ECG channel recording) must be available in a Matlab variable. For the purpose of this manual, this variable is called data.
Reference (help)
Output of help heartbeats:
heartbeats - detect heart beats and frequency in physio data
FORMAT: [bp, bs, bf, bv, cp, wgd, wd] = heartbeats(sig [, opts])
Input fields:
sig Sx1 numeric signal
opts optional settings
.bppre pre-defined positions (no detection, only inspection)
.cleanup interactive cleanup (default: false)
.detlength detection length threshold in seconds (default: 0.05)
.pflength pre-filter length in seconds (default: 0.025)
.pfreps pre-filter repetitions (default: 2)
.freq data frequency in Hz (default: 1000)
.plot plot mean +/- std estimate of signal (default: false)
.segsize segmentation size in seconds (default: 5)
.segstep stepping (window shift) in seconds (default: 1)
.windsor windsorizing threshold in std's (default: 3)
Output fields:
bp beat positions
bs beat positions (in seconds)
bf frequency estimate for each beat
bv values at peak
cp estimate of centers of windows (one value less!)
wgd guess whether window is good or not
wd windowed data (in 100Hz resolution, interpolated)
Note: this function is still preliminary
Usage overview
Once the data is loaded, the function call is relatively straight forward. For the “first pass”, cleanup should be enabled, as well as plotting the results:
[bp, bs, bf, bv, cp, wgd, wd] = ... heartbeats(data, struct('cleanup', true, 'plot', true));
This will perform the following steps:
- windowed z-transform (in segment windows with configured stepsize, also works as low-pass filter!)
- windsorizing (cutting off over-large peaks)
- filtering (low-pass, to get rid of very high frequency noise)
- skew-flipping (so that peaks always are on the positive tail of the distribution)
- segment-wise beat detection (maximum value position within each sub-segment where value > mean + 0.5 * skew)
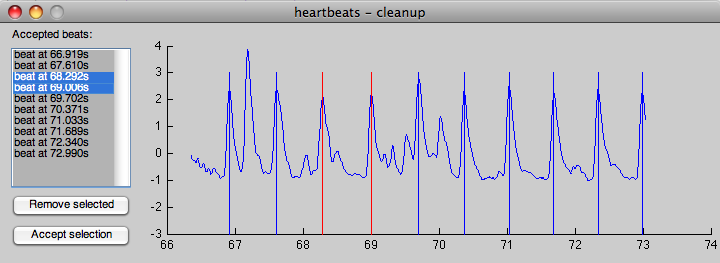
Next, the detected (or provided) beats will be displayed in a small dialog for places where either the distance between beats is too small or too large (and the shape of the detected beat does not match the mean signature):
The operation is relatively simple:
- to add a beat in the stream, simply press the mouse button close to the desired peak location (which will be determined in a +/- 100ms window around the clicked spot)
- to remove a beat (or several beats), select those in the dropdown box and press the “Remove selected” button
- once all beats in this window are correct, press the accept button
If no more bad beats are in the list, the dialog will close automatically. If you wish, you can also manually close the dialog (prematurely) to go on to the next stage.
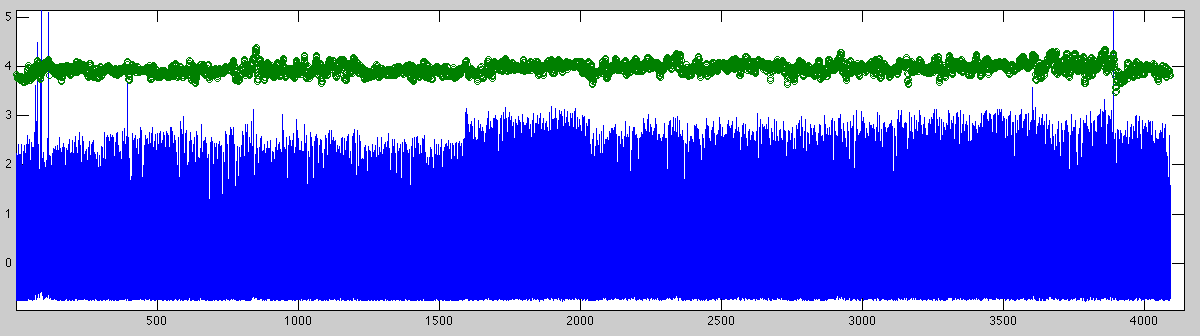
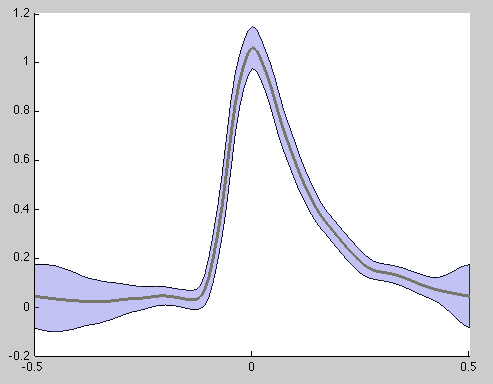
In case the plotting has been enabled, the following two displays appear:
and
The second figure simply shows a representation of the mean (each window is re-sampled to match a 100-sample window length), whereas the X-axes is labeled as phase with the peak being located at 0.
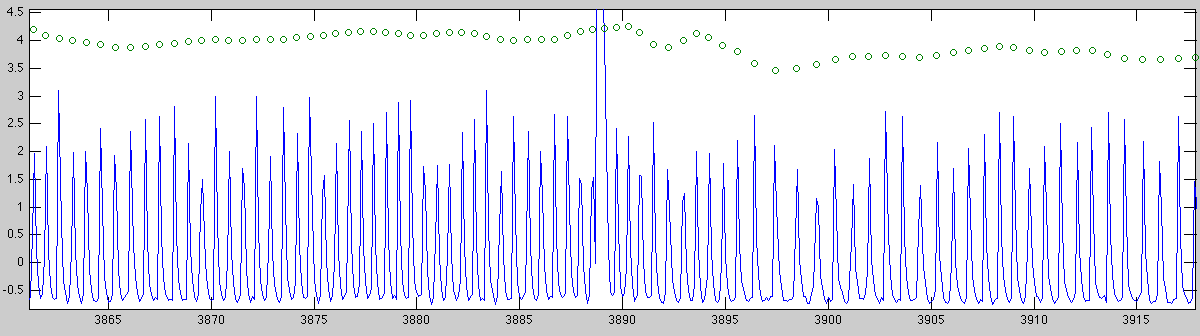
The first figure shows the entire signal over time in a 20Hz sampling (in blue) with the detected heart-rate super-imposed to control for flaws in the detection. This can be done by zooming in on a smaller piece of the signal: