Table of Contents
Flatmaps
Motivation
While surface projection of results already makes a much larger amount of cortical activation visible “at a glance” (compared to showing single slices, for instance), it still requires at least two separate images for the lateral and medial portion of cortex (for one hemisphere).
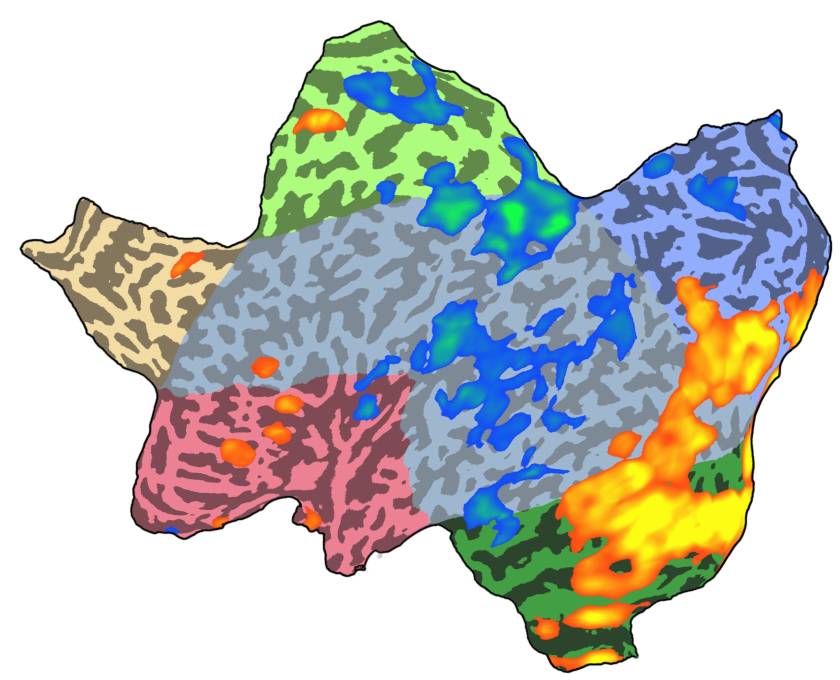
One way to circumvent this problem is by producing what people in the field have coined “flatmaps”. As part of the processing, the surfaces are first hugely inflated, then “cut open” (kind of like the peel of an orange; usually with a small patch of surface being removed entirely to minimize later distortion) followed by flattening the remaining vertices into a two-dimensional plane (which usually includes some kind of distortion correction, given the high amount of deformation this procedure introduces).
The result usually looks something like this:
Requirements
To create flatmaps such as the one shown above you need
- a statistical map you want to visualize (can already be in SMP format, otherwise VMP)
- the surface file representing the folded hemisphere (structure, used to sample statistical or functional data)
- a flattened version of the surface file with identical number of vertices and neighborhood information (vertex inter-connectivity)
In case you do not have a surface file of your own, you can use the surface files created during a call to neuroelf_makefiles which are a representation of the single-subject brain used in SPM, Colin_27 (the 163,842-vertices version of the left hemisphere of this dataset was used to create the above screenshot).
Procedure
Two paths are described below: sampling existing statistical information (in VMP format) at the spatial location of folded cortex and displaying it as a flatmap or sampling timecourse information and creating statistics (currently only implemented in BrainVoyager QX!) and then using the required SRF and SMP objects from BrainVoyager in NeuroElf for a high-quality screenshot.
VMP-based
If you have a VMP with one (or sevaral) map(s) you wish to visualize as a flatmap
- load and select the VMP in NeuroElf GUI (either by using the File → Open file request dialog or by issuing
vmp.Browse;on the VMP object variable) - load and select the desired folded surface you wish to use for sampling the statistical data into surface-space (again either by using File → Open or
srf.Browse;) - select the
Create SMP…entry from the VMP menu - optionally store the created SMP (e.g. for later use in NeuroElf or BrainVoyager QX)
- ensure that the shown map is correctly displayed (activations should be on the correct hemisphere and in the right orientation!)
- load and select the corresponding flattened surface (which must share the number of vertices and neighborhood information!)
- if required, select the newly created SMP for this flattened surface, then select the desired map (if multiple maps are available)
- click the “undock” button of the main UI (top button in the View/Page selection toolbar)
- use the SHIFT + S keyboard command to invoke the screenshot function
- select Encapsulated PostScript (*.eps) as file format
- save the file
- open (or switch to) a graphics editing program (such as Adobe Photoshop or The GIMP)
- open the saved file and select a high-enough resolution (at least 300DPI) and the desired color space (RGB or CMYK) for rendering
- optionally re-touch the image (for the screenshot above, the black background was replaced with a white background, leaving a small black fringe/border using the magic-wand-tool)
- save the image to a different format (rendered, bitmap)
MTC/SRF-based
This is only required in case you wish to use NeuroElf for saving surface screenshots in EPS format!
- use BrainVoyager to compute the statistics in SMP format (see BrainVoyager manuals for help)
- save the SMP
- load the flattened surface (SRF) and corresponding SMP in NeuroElf
- continue with the above list of instructions starting with the “undock” button instruction